CNN Cardiac Rhythm Classification
Problem Statement
The MIT-BIH Arrhythmia dataset contains annotated ECG heartbeats from a wide range of arrhythmia types. While traditional ML methods perform well on smaller ECG datasets like ECG200 and ECG5000, they fail to generalize to the more complex MIT-BIH dataset, particularly when using simple feed-forward MLP architectures. The goal of this project is to build a robust 1D CNN model capable of accurate multi-class heartbeat classification on MIT-BIH data.
Theoretical Approach
After my attempts to classify this dataset with a deep resnet MLP demonstrated suboptimal performance, I suspected a more complex model was needed. CNNs excel at image classification, so may be more appropriate for the task. As my previous work involved creating networks from scratch, I used this project as an opportunity to learn more about Pytorch and Google Colab.
A residual convolutional neural network was trained on single lead (II) ECG tracings from the public dataset available on Kaggle. There was significant class imbalance with most being of the normal class. To address significant class imbalance, both weighted sampling and focal loss were employed. Noise injection during training provides mild regularization.
Purpose
This project is my first full implementation using PyTorch and Google Colab. I used ChatGPT and online resources extensively to help learn syntax, explore PyTorch architecture patterns, and debug training errors. The objective was to go beyond basic MLPs by implementing a CNN capable of learning spatial features directly from the ECG waveform. The project serves as a learning exercise and proof of concept that a CNN is better suited to classify MIT-BIH data than a MLP.
Methods
- Data: MIT-BIH heartbeat dataset downloaded via Kaggle API and preprocessed in Google Colab
- Classes: Class label representation was as follows:
- 0 : Normal
- 1 : Supraventricular premature beat
- 2 : Premature ventricular contraction
- 3 : Fusion of ventricular and normal
- 4 : Unclassifiable beat or noise
- Normalization: Each ECG beat was standardized individually
- Model: DeepECGResNet1D, with 4 residual blocks and adaptive average pooling
- Loss: Focal loss to handle class imbalance
- Sampling: Weighted random sampler
- Training: Early stopping and learning rate scheduler
- Visualization: Loss curves, filter visualization, and confusion matrix
Results
- Final validation accuracy: ~95%
- Most classification errors on Supraventricular and Fusion beats classified as Normal
- Balanced precision and recall across most classes, especially after applying weighted sampling and focal loss
precision recall f1-score support
0 0.9955 0.9473 0.9708 18118
1 0.4464 0.8759 0.5914 556
2 0.8867 0.9517 0.9181 1448
3 0.4573 0.9259 0.6122 162
4 0.9535 0.9944 0.9735 1608
accuracy 0.9491 21892
macro avg 0.7479 0.9390 0.8132 21892
weighted avg 0.9673 0.9491 0.9552 21892
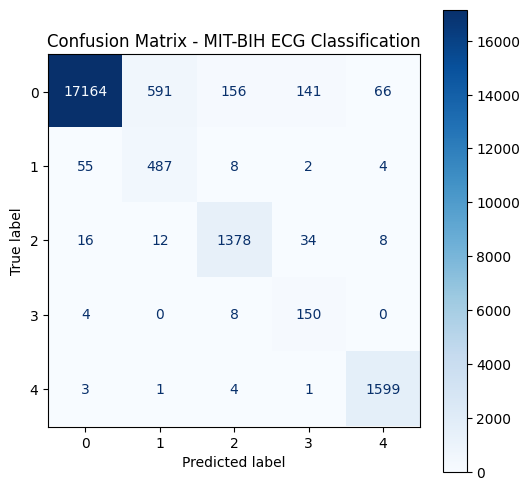
Conclusion
This project demonstrates the effectiveness of deep residual CNNs for time-series classification on ECG data. By combining PyTorch tools like WeightedRandomSampler, FocalLoss, and modular design, this model significantly outperforms earlier MLP approaches on real-world ECG data.
Future Work
This work may be extended by train a model on multi-lead ECG data classification
References
No external publications were used, though LLM queries aided architecture and debugging.